Fanzor proteins show potential for precise editing of the human genome, offering an alternative to CRISPR-Cas systems.
Scientists from the McGovern Institute for Brain Research at MIT and the Broad Institute at MIT and Harvard have made a significant discovery Fanzor. While the name might sound like it belongs to a character expected to battle Godzilla, Fanzor proteins play a key role in a new RNA-guided genome editing system, and one that opens up new possibilities for more DNA-editing. accurate in human cells compared to the widely used CRISPR/Cas systems.
Posted in Naturethis new study provides insights into the structure and function of Fanzor proteins, highlighting their potential as a therapy for human genome editing.
Longevity. Technology: CRISPR-Cas systems, originally identified in prokaryotes such as bacteria, have revolutionized genetic research due to their ability to be easily reprogrammed to target different sites in the genome. However, scientists have long speculated that similar systems exist in more complex eukaryotes, the domain that includes fungi, plants and animals. Researchers have now demonstrated the presence of RNA-guided DNA cutting mechanisms in all kingdoms of life, establishing Fanzor as a promising alternative to existing genome-editing tools.
CRISPR, short for Clustered Regularly Interspaced Short Palindromic Repeats, is a powerful gene editing technology that allows scientists to make precise changes in the DNA of organisms by using a molecule called RNA as a guide to target specific DNA sequences and make changes in those locations . Fanzor compact systems introduce a new RNA-guided system that can be more easily upgraded to improve the precision and delivery of genome editing therapies.
CRISPR-based systems are widely used and powerful because they can be easily reprogrammed to target different sites in the genome, said Zhang, the study’s senior author and principal institute fellow at Broad, a research scientist at MIT’s McGovern Institute, the James and Patricia Poitras Professor of Neuroscience at MIT and researcher at the Howard Hughes Medical Institute. This new system is another way to make precise changes in human cells, complementing the genome editing tools we already have [1].
The researchers isolated Fanzor proteins from various organisms, including fungi, algae, amoeba species and a clam called a northern quahog. Through meticulous biochemical characterization, they discovered that Fanzors are endonuclease enzymes that use non-coding RNAs known as RNA to precisely target specific sites in the genome [2]. This mechanism, first observed in eukaryotes, showcases the versatility and potential of Fanzor proteins for genome-editing applications in animals.
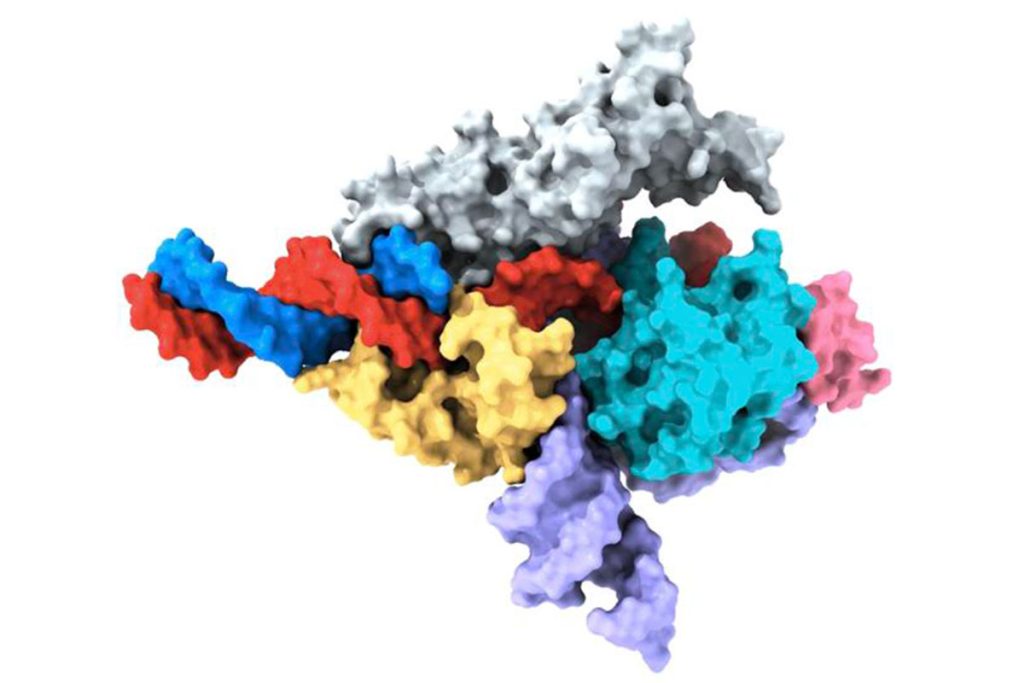
Photograph: Zhang Lab, Broad Institute of MIT and Harvard/McGovern Institute for Brain Research at MIT
Unlike CRISPR proteins, Fanzor enzymes are encoded within the transposable elements of the eukaryotic genome. Phylogenetic analysis indicates that Fanzor genes migrated from bacteria to eukaryotes via horizontal gene transfer.
This is also strengthened by previous research; two years ago, members of the Zhang lab discovered a class of RNA-programmable systems in prokaryotes called OMEGAs, which are often linked to transposable elements, or jumping genes, in bacterial genomes and likely gave rise to CRISPR/Cas systems. That work also highlighted similarities between prokaryotic OMEGA systems and Fanzor proteins in eukaryotes, suggesting that Fanzor enzymes might also use an RNA-driven mechanism to target and cut DNA.
These OMEGA systems are more ancestral than CRISPR and are among the most abundant proteins on the planet, so it makes sense that they would have been able to jump back and forth between prokaryotes and eukaryotes, explained co-first author Makoto Saito of the Zhang lab. [1].
To evaluate Fanzor’s editing capabilities, the researchers successfully demonstrated its ability to generate insertions and deletions at targeted genome sites in human cells. While initially less efficient than CRISPR-Cas systems, the team engineered the Fanzor protein through a combination of mutations, resulting in a tenfold increase in activity. Importantly, unlike some CRISPR systems, the Fanzor proteins showed minimal ‘side activity’, mitigating the unintended cleavage of nearby DNA or RNA.
Structural analysis conducted by the team revealed similarities between Fanzor and its prokaryotic counterpart, the CRISPR-Cas12 protein. However, the interaction between RNA and Fanzor’s catalytic domains was more extensive, suggesting the involvement of RNA in catalytic reactions. This insight into Fanzor’s molecular structure provides a basis for further design and optimization to improve its accuracy and efficiency as a genome editing tool.
We are excited about these structural insights for helping us further design and optimize Fanzor for greater efficiency and accuracy as a genome editor, said co-first author Peiyu Xu [1].
Fanzor shares the defining characteristic of CRISPR-based systems, its programmability to target specific sites in the genome. This feature, combined with the potential for increased efficiency and accuracy through ongoing research, positions Fanzor as a promising technology for both research and therapeutic applications in the future. Furthermore, the prevalence of RNA-guided endonucleases such as Fanzor suggests the possibility of discovering novel RNA-programmable systems, expanding our understanding of genetic mechanisms in diverse organisms.
Fanzor’s discovery marks a significant milestone in the field of genome editing, offering scientists a potential alternative to CRISPR-based systems for precise manipulation of the human genome.
Nature is amazing. There is so much diversity, Zhang said. There are probably more RNA-programmable systems out there, and we’re continuing to explore and hopefully find out more. [1].
[1] https://news.mit.edu/2023/fanzor-system-in-animals-can-edit-human-genome-0628
[2] https://www.nature.com/articles/s41586-023-06356-2
Photography: vectorpocket/Freepik
#Researchers #discover #RNAguided #genome #editing #system
Image Source : longevity.technology
