
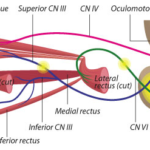
Elizabeth Engle, MD, has dedicated her career to researching genetic and developmental causes for eye, lid, and facial movement disorders. From common conditions like strabismus to very rare ailments, these conditions can affect a person’s appearance and impair social communication, making it difficult to move the eyes up, down or to the side, or regulate facial expressions.
Every discovery in Engles’ lab offers an answer for the families. It is also a window to the brain, particularly to the development of the nerves and motor neurons that innervate the muscles of the eyes and face.
Now, after nearly a decade, Engles’ team at Boston Childrens Hospital’s FM Kirby Neurobiology Center has unraveled a previously impenetrable mystery: the genetic cause of a condition called hereditary congenital facial paresis type 1 (HCFP1).
HCFP1 causes mild facial weakness that makes it difficult for infants and toddlers to smile brightly, pucker their lips, suckle, and sip from straws. Though mild and very rare, the condition has opened the door to a poorly understood area of genetics: disorders caused by inherited mutations in the dark matter of the genome.
A rare category of genetic disease
Most of the known genetic causes of inherited diseases result from variations in genes in the 1 to 2 percent of our genome, also known as the exome, which provides the code for making proteins. But within the rest of the genome, dark matter contains important non-coding elements that regulate genes and can influence disease.
Although we only have about 20,000 genes, noncoding regulatory elements provide a lot of additional information, modulating gene expression by turning genes on or off in specific cell types at specific times, says Engle.
Single-gene inherited disorders involving noncoding variations remain largely underexplored, in part because we do not yet know how to interpret most variations within the vast noncoding space. The new work provides a rare example that could inform future genetic discovery for other disorders.
Examine the genetic variants of families
The team started with nine families from the Engle laboratory database and five families of collaborators in the Netherlands. Linkage mapping in Dutch families had identified the general area in the genome that appeared to be linked to HCFP1. The families had had whole exome sequencing, but since this only includes the coding parts of the genome, it came up blank.
To solve questions like this, you need a team of people with multiple areas of expertise. We think our work provides an example of making fun of the genetics and mechanisms of inherited diseases involving non-coding variations.”
Engles’ team used whole genome sequencing, which revealed that five families had a large piece of duplicated DNA. The starting and ending points of this piece varied, but in all five families it contained three non-coding normative elements. The other nine families had single-letter single-nucleotide variants (SNVs) in the genetic code, all within one of the three regulatory elements.
This is a bit of a conundrum, because we typically think of DNA duplications as an increase in gene expression and SNVs as a decrease in gene function, says Engle. We had to understand the mechanism by which these duplications and SNVs produced the same facial phenotype and determine which gene these regulatory elements were targeting.
They’ve discovered that the three Regulators are nearby GATA2, a gene known to help specify the production of different cell types in the blood and nervous system. This begged the question: are the three regulators taking action GATA2? Engles’ team suspected he had, and that two augmented and one silenced GATA2 expression.
Tracking the developmental journeys of cells
This is where a lot of footwork came into play. Using mouse models and tools such as single-cell RNA sequencing and CUT&Tag to profile cells at different time points, the team has begun to understand how variants in DNA regulatory elements lead to facial weakness.
First, they found that regulatory elements are active during early brain development, at a location in the hindbrain where two types of cells arise: facial motor neurons and inner ear efferent neurons (involved in filtering loud and loud sounds). background noise).
Our hypothesis was that the inner ear effervescents arise earlier and depend on the activation of the stimulators GATA2and that about a day later the muffler goes off GATA2 and facial motor neurons are done, says Engle. We thought that DNA duplications could increase enhancer activity while SNVs could decrease silencing activity, which would both lead to the production of more inner ear efferents and fewer facial motor neurons.
Meticulous experiments have shown this to be true. The combined number of cells was unchanged, but extended GATA2 the expression increased inner ear efferents at the expense of facial motor neurons, says Alan Tenney, PhD, one of the postdoctoral researchers in Engles’ lab who led the study.
An explanation for HCFP1
In summary, the team showed that genetic mutations prevent people from producing enough motor neurons during development to adequately innervate facial muscles, leaving them relatively weak.
To solve questions like this, you need a team of people with multiple complementary areas of expertise, says Engle. We think our work provides an example of mocking the genetics and mechanisms of inherited diseases involving non-coding variations. The key is what you choose to address which noncoding variants are compelling enough to spend the next five years of your life investigating.
Tenney, Alessandro Di Gioia, PhD, and Bryn Webb, MD, were co-first authors of the study, published in Genetics of nature.
Learn more about the research in the Engle lab and the FM Kirby Neurobiology Center at Boston Childrens.
Related posts:
-

Tracing the elusive genes that cause strabismus
Strabismus is a common condition in which the eyes don’t line up properly, rolling in, out, up, or down. Two …
-
The neurology resident who could
Elizabeth Engle used to wait in people’s driveways until midnight, hoping to enroll them in her genetic eye movement studies…
-

Two neuroscience rock stars elected to the National Academy of Medicine
Beth Stevens, PhD, and Elizabeth Engle, MD, are the latest Boston Childrens Hospital researchers to be elected to the…
-
The beauty of the brain
Each year, the Harvard Brain Science Initiative sponsors the Beauty of the Brain contest. This year, two Boston Childrens…
#Facial #Weakness #Dark #Matter #Detective #Story #Boston #Childrens #Answers
Image Source : answers.childrenshospital.org
